April 14, 2020
Faculty of Science researchers to design new drug leads, inspired by existing anti-viral drugs

Understanding the anatomy of viruses, and repurposing the chemical compounds found in drugs to treat them, may help researchers get one step closer to quickly developing effective medication for the COVID-19 virus.
By proposing to develop more effective antiviral chemical compounds to help form the bases of new drugs, two researchers in the Faculty of Science have received federal recognition and support for a molecular-level discovery research project. Dr. Kenneth Ng, PhD, from the Department of Biological Sciences, and Dr. Chang-Chun Ling, PhD, from the Department of Chemistry, received a $416,000 operating grant from their application to the Canadian Institutes of Health Research (CIHR) Canadian 2019 Novel Coronavirus (COVID-19) Rapid Research Funding Opportunity competition.
- Photo above: Model COVID-19 polymerase enzyme bound to RNA and proposed inhibitor inspired by models of Remdesivir.
Their multidisciplinary work builds on promising initial results of the drug Remdesivir, which was developed originally to combat Ebola virus infections and is currently being tested in clinical trials in China and the United States for treating COVID-19. If successful, their work on chemical compounds to target the coronavirus could form the foundation of new and more effective drug therapies for COVID-19.
Collaboration helps pivot existing work to quickly address new disease
Ng and Ling have worked on projects together for more than 15 years as investigators in the Alberta Glycomics Centre and the Canadian Glycomics Network. Fortunately, this long-standing research partnership allowed them to quickly pivot some of their existing work to rise to the challenge of treating COVID-19.
To meet CIHR’s deadline, “It was a very fast turnaround to write this proposal — mostly over the Family Day long weekend,” says Ng, who has been working on understanding the polymerase enzyme to develop treatments for other viral diseases for nearly 20 years.
“I knew Dr. Ling had the complementary expertise in organic chemistry to translate our knowledge of the molecular structure into new lead compounds for possible treatments. Fortunately, he could see how he could help and agreed right away to join the project.”
The coronavirus causing COVID-19 relies on an enzyme called the nsp12 polymerase to make copies of its RNA genome to make new viral particles during infection. “Our 3D models of nsp12 bound to RNA show how existing drug compounds developed to treat other viral diseases could be modified to work more effectively for treating COVID-19,” Ng explains.
“Based on this analysis, Dr. Ling’s lab will synthesize a series of modified chemical compounds tailored to fit the structure of nsp12, and my lab will test the effects of these compounds on the purified enzyme. Our team will also determine how nsp12 recognizes these compounds in 3D so that we can improve the design of the next generation of compounds to work even more effectively.”
By disrupting the critical function of this enzyme in the production of new virus particles, these new compounds should help patients to overcome the disease, which is especially important in severe cases.
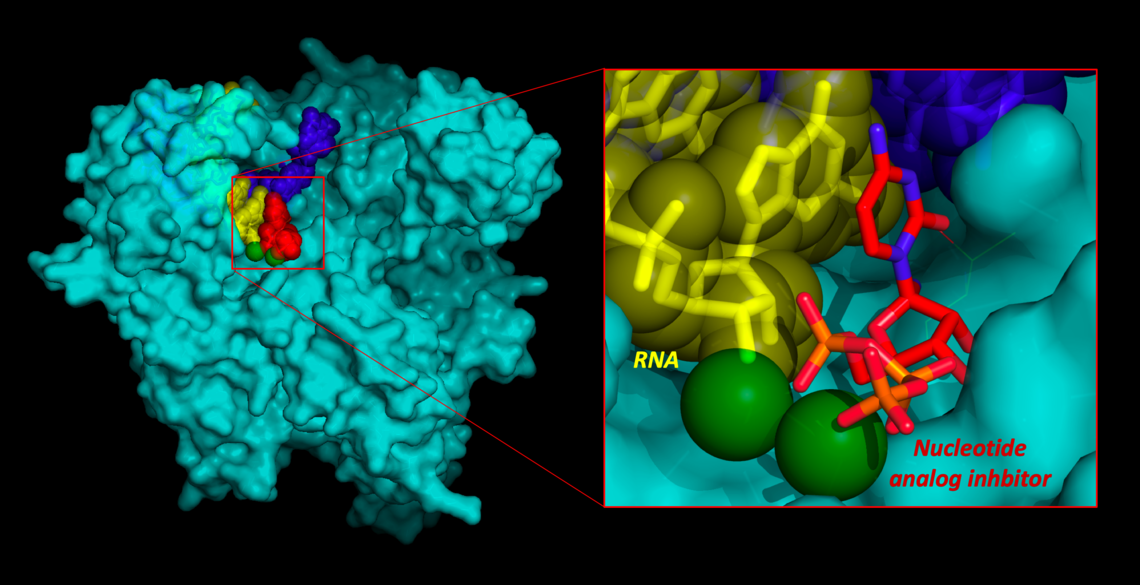
Model COVID-19 polymerase enzyme bound to RNA & proposed inhibitor inspired by models of Remdesivir.
Kenneth Ng
Developing highly specific drugs at the forefront of COVID-19 fight
While humans, plants, and animals all have DNA (deoxyribonucleic acid) as biological building blocks, many viruses use RNA (ribonucleic acid) — a genetic material that is nearly identical to DNA, but with one important fundamental difference. Ng and Ling plan to exploit this difference, using drugs specific for the viral RNA-dependent RNA polymerase to prevent the virus from reproducing.
Taking inspiration for their work from the drug Remdesivir was a deliberate choice, and one that Ling says may help to accelerate the process of creating new and more effective treatments. Initial efforts toward drug treatments for COVID-19 are building on existing drugs because these have already gone through the necessary validation, quality assurance and trials to rule out detrimental side effects.
“Many viruses, including Ebola and the flu, have an RNA genome,” says Ng. “The polymerase enzyme that copies the RNA genome is therefore different from similar human enzymes that instead work on a DNA genome. This difference gives us a much better chance to design a drug that is very specific against the virus so it doesn’t have the negative effects of interfering with important human enzymes.
Our goal is to make something that targets the coronavirus very specifically, with minimal negative side effects.
They hope that the compounds with the strongest effects on this critical enzyme can be used at low doses to minimize side effects while providing maximum benefit for the most severely affected patients.
“We’re hoping that the modified compounds we’re aiming to synthesize will match specific features of the COVID-19 RNA polymerase like a key in a lock and give us that selectivity,” Ling explains.
Collaboration and support to help deliver results
The teams expect to prepare and test the first series of compounds within four to six months. The 3D structural analysis will take another four to six months, followed by a second round of compound synthesis and testing. The most promising compounds from the first and second rounds of synthesis will be passed on to collaborators and the pharmaceutical industry to evaluate their effectiveness in cell culture and animal infection models, as well as human clinical trials.
The process is highly collaborative. “What we’re trying to do is to super-accelerate the generation of data on a new series of compounds. The complex process of drug development is a very collaborative, multi-step process where we will do the early part of discovering active enzyme inhibitors, and then others will take it further to see which ones are not toxic in people but also very effective against the virus,” Ng says.
Support for this begins at their home institution, where Ling says UCalgary is “really making an effort to assist us.
“The associate dean of research, department head, and infrastructure manager have been very helpful in helping get access to labs and supplies. Without their support, it would not be possible.”
Ng says the whole spectrum of drug and vaccine development is very important. “It’s going to take a while to get a good vaccine, which will eventually be the most important defence against the virus. While good vaccines are in development and even after they are available, there will also be a need to have better drugs, or more drugs available, to deal with cases of severe illness.
"RNA viruses can develop resistance to drugs very quickly. That’s a universal fact about any virus or infectious disease, because the organism is always trying to find a way to get around the drug. I think that’s why our work to create new options for treating the disease is especially valuable.”
Says Ling, “The severity of this current situation is really requiring us to find solutions using different approaches to see which one works best. Focusing on the fundamental science allows us to build an arsenal to fight viruses in future situations.”
COVID-19 resources
- For the most up-to-date information about the University of Calgary's response to the spread of COVID-19, visit the UCalgary COVID-19 Response website.
- For resources to support students, faculty, staff, alumni, and all our communities during this unprecedented time, visit the UCalgary COVID-19 Community Support website.
- Go to the Campus Community Needs Assessment page if you need assistance with basic needs, or physical or mental health supports.
- Register with the COVID-19 Volunteer Support page if you are a UCalgary staff, faculty, student or alum who has the volunteer capacity to help; we will match you with a member of our community in need.





